These publications are a subset of my scientific work that I consider to have contributed significantly towards. For further auxiliary publications see my Google Scholar.
2025
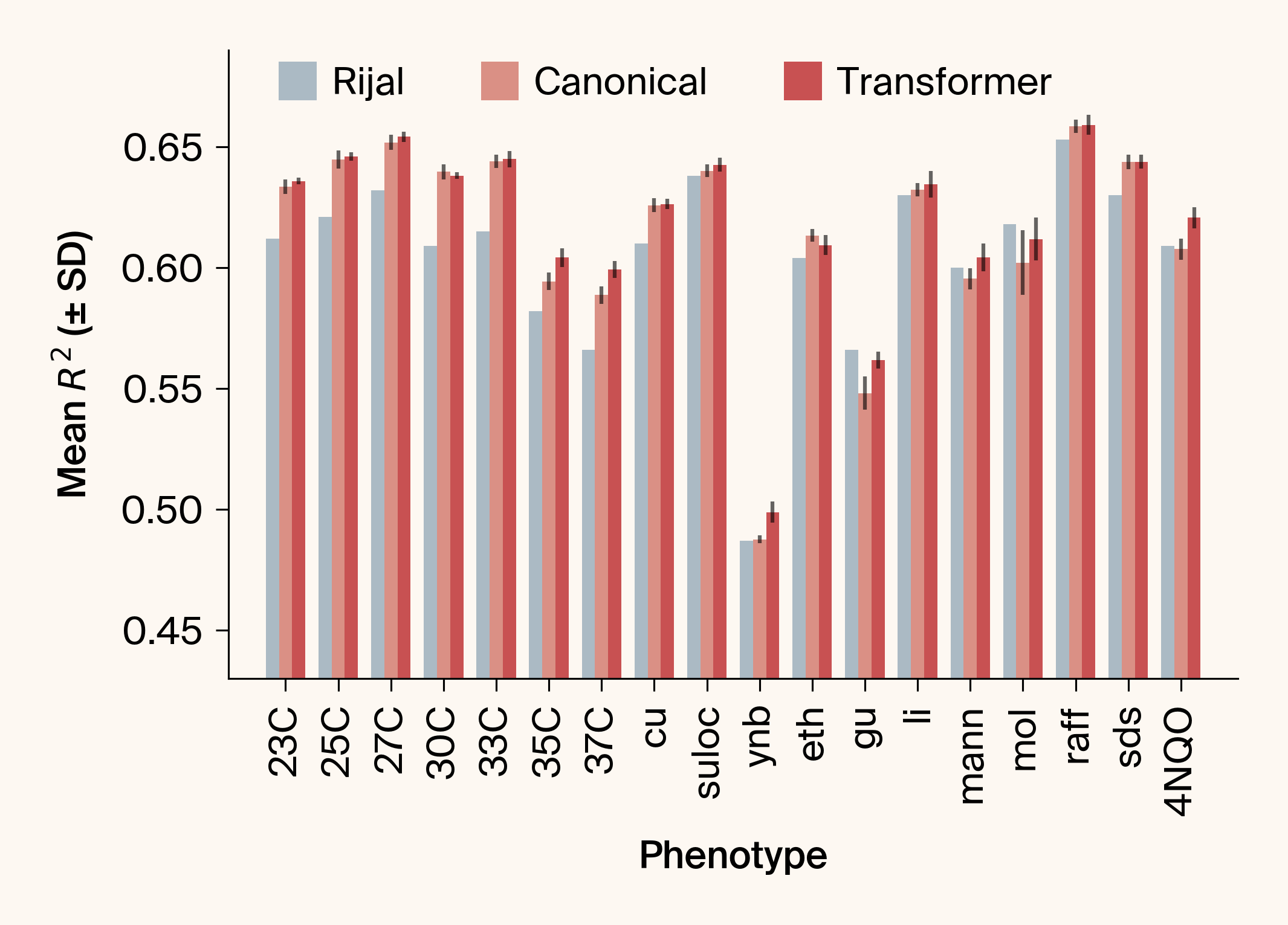
- Asks: can standard transformers outperform Rijal et al.'s custom attention architecture for genotype-phenotype mapping?
- Using a large genetic yeast dataset, we found canonical ML components significantly outperformed the bespoke design
- Key insight: multi-objective training exploits cross-phenotype genetic correlations, allowing the network to leverage mutual information between related traits for improved prediction
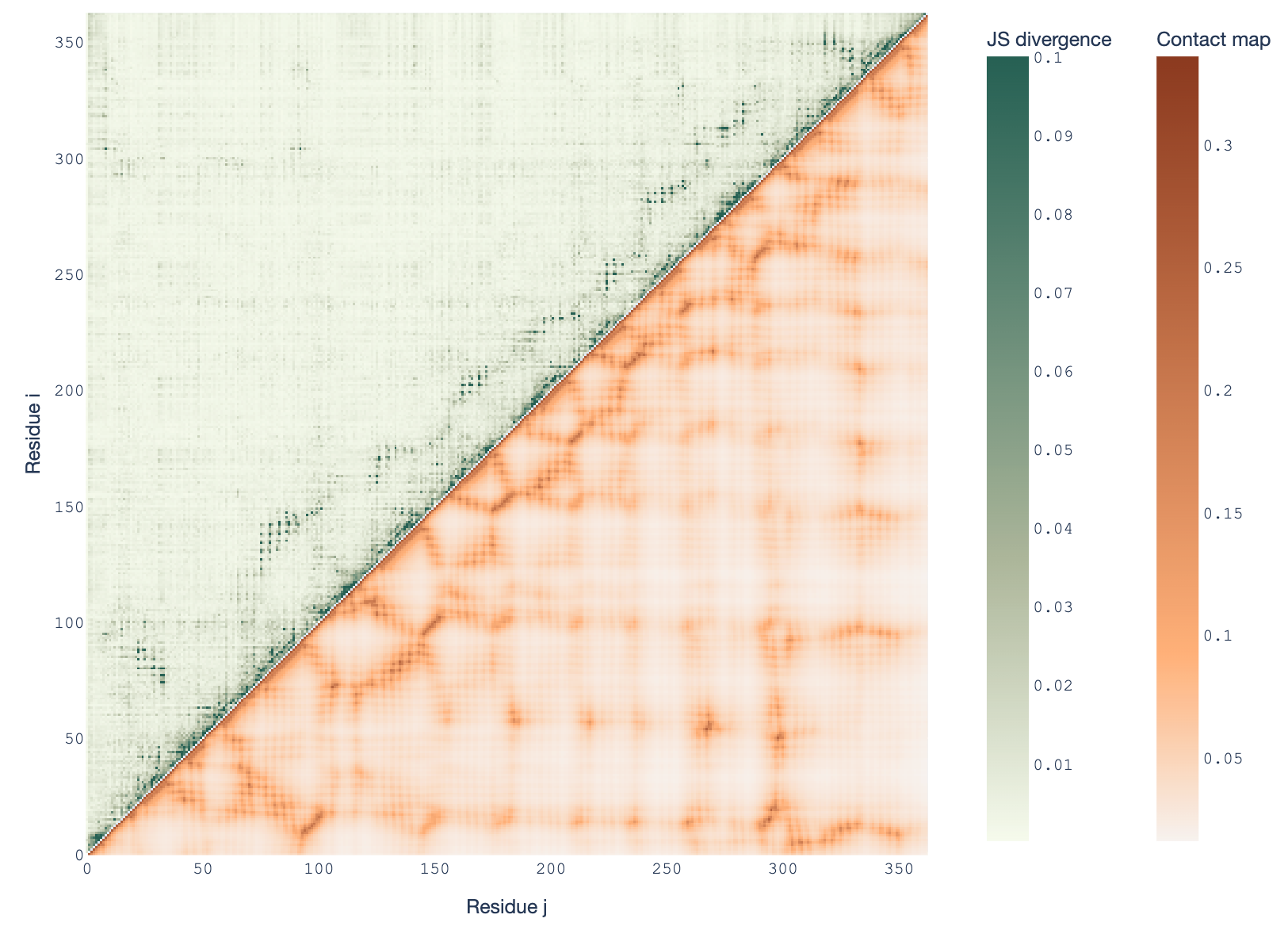
- We discovered an unexpected pattern in the ESM2 protein language model family: amino acid probability distributions mirror protein 3D contact maps, but this weakens in larger models.
- Using Jensen-Shannon divergence, we found intermediate models show the strongest correlation with structural contact maps, leaving the largest models in the dust.
- This counterintuitive result challenges assumptions about model capacity and biological interpretability.
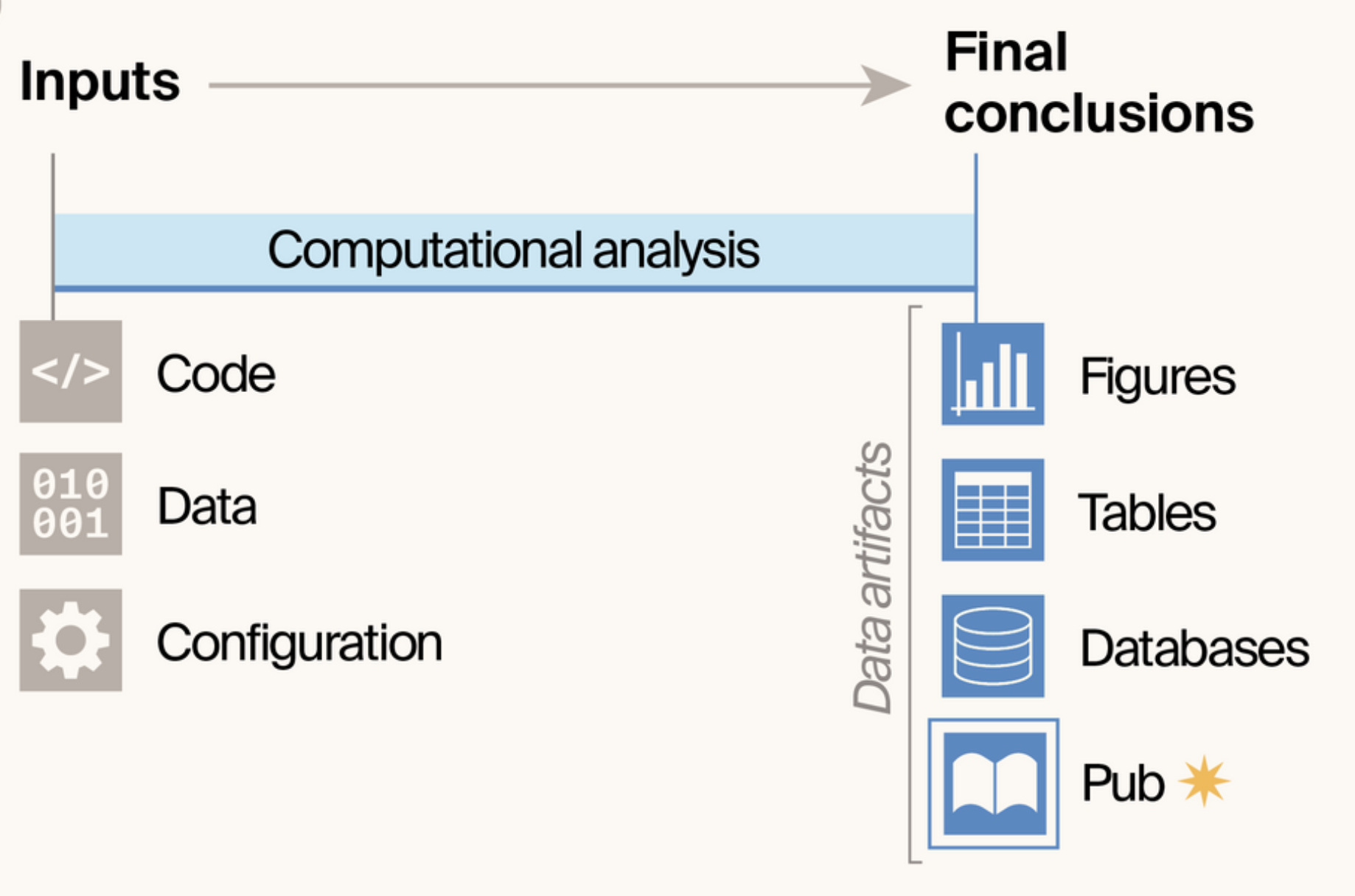
- Introduces notebook pubs: a publishing format that treats computational notebooks as publications themselves, eliminating the gap between how scientists analyze data and how they share results.
- By making the publication itself a data artifact of the analysis pipeline, notebook pubs ensure end-to-end reproducibility while reducing publication burden and enabling faster sharing of results.
- Built on Quarto and GitHub infrastructure, the approach provides scientists with a template that automatically converts Jupyter Notebooks into hosted, interactive web publications with version control and community engagement built in.
2024
- A general-purpose billiards simulator designed specifically for science and engineering applications with a focus on speed, ease of visualization, and fine-grained analysis.
- Features an event-based simulation algorithm with JIT compilation that significantly increases computational efficiency compared to traditional time-step methods, by precisely calculating when significant events like collisions occur.
- Provides an interactive 3D interface with comprehensive playback controls and a controllable camera for visualizing shot trajectories in a realistic environment.
- Bridges a critical gap in billiards research by offering an open-source platform with realistic physics that can be used across disciplines including game theory, robotics, computer vision, and sports analytics.
2023
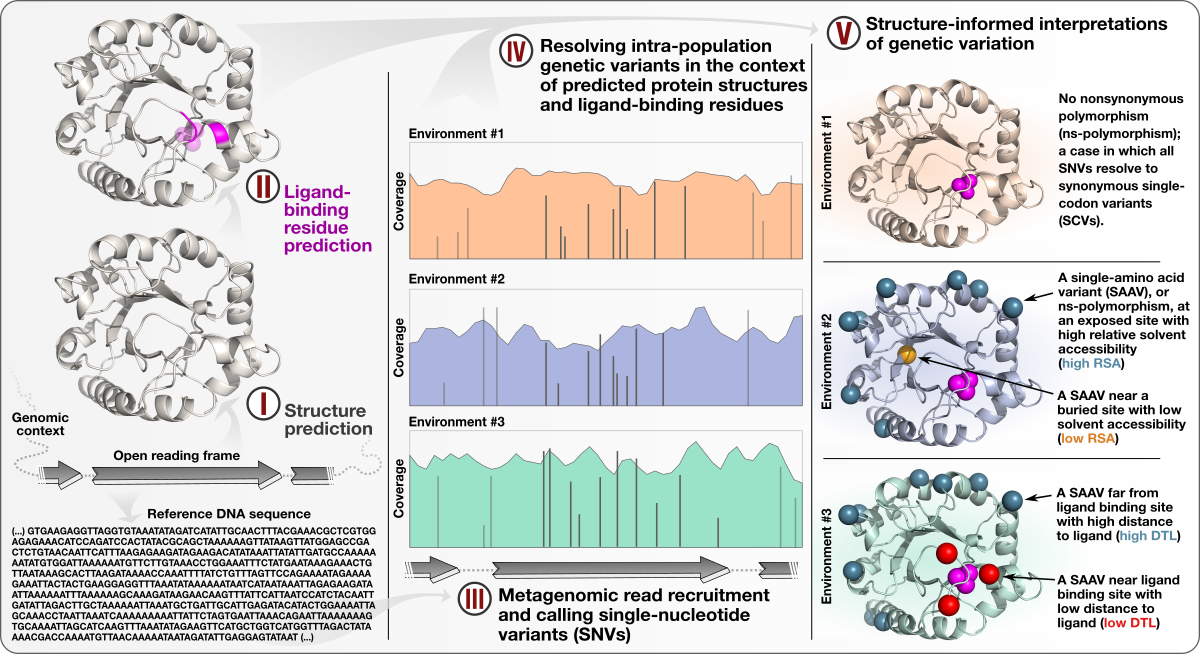
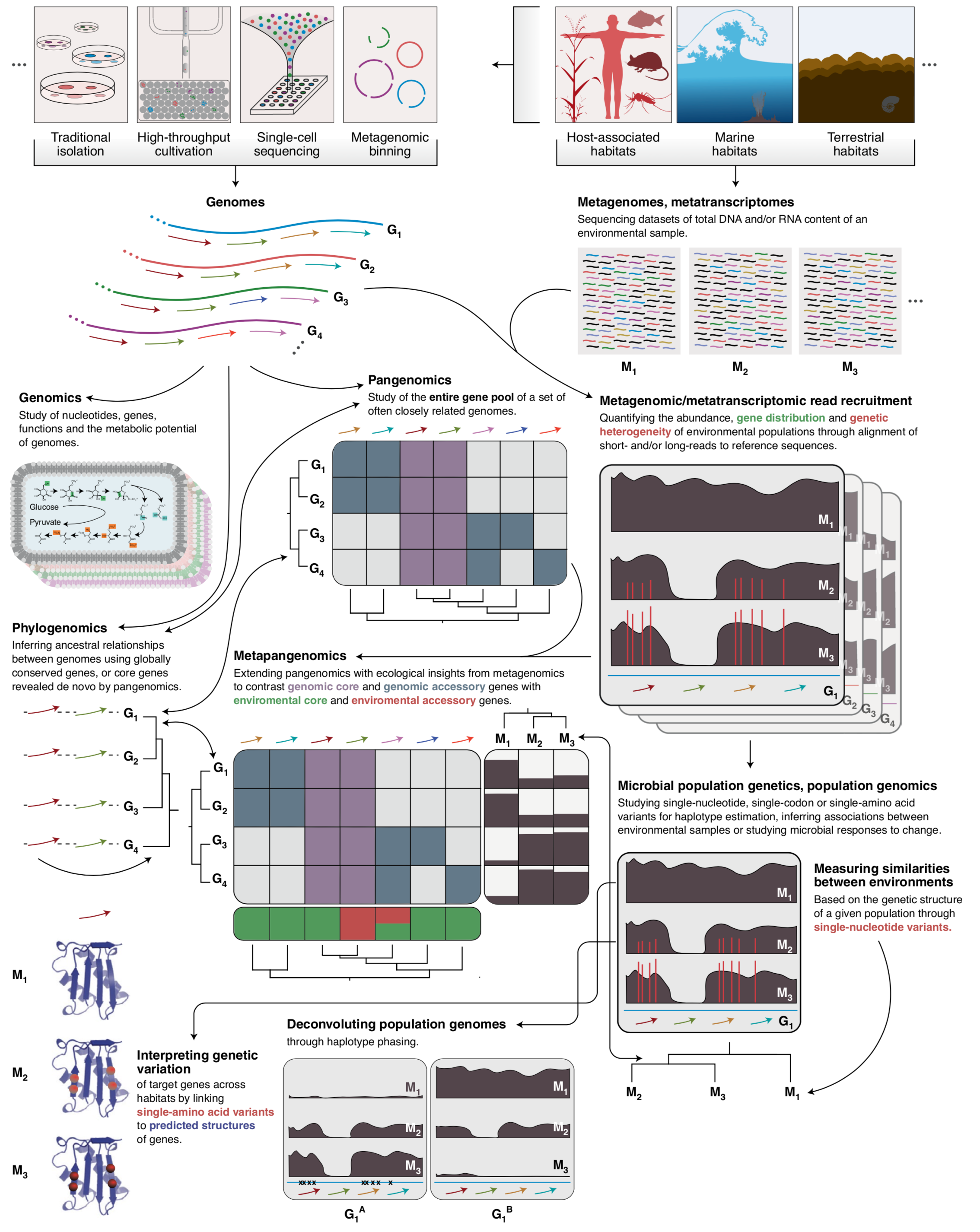
- A study that describes an approach to integrate environmental microbiology with recent advances in protein structure prediction, and illustrates the tight association between intra-population genetic variants, environmental selective pressures, and structural properties of proteins
- Demonstrates a quantifiable link between (1) the magnitude of selective pressures over key metabolic genes (e.g., glutamine synthase of the central nitrogen metabolism), (2) the availability of key nutrients in the environment (e.g., nitrate), and (3) the maintenance of nonsynonymous variants near protein active sites.
- Shows that the interplay between selective pressures and protein structures also maintains synonymous variants -- revealing a quantifiable link between translational accuracy and fluctuating selective pressures.
- Comes with a reproducible bioinformatics workflow that offers detailed access to computational steps used in the study that spans from metagenomic read recruitment and profiling to the integration of environmental variants and predicted protein structures.
2020
- A summary of the progress of anvi'o during the past five years.
2019
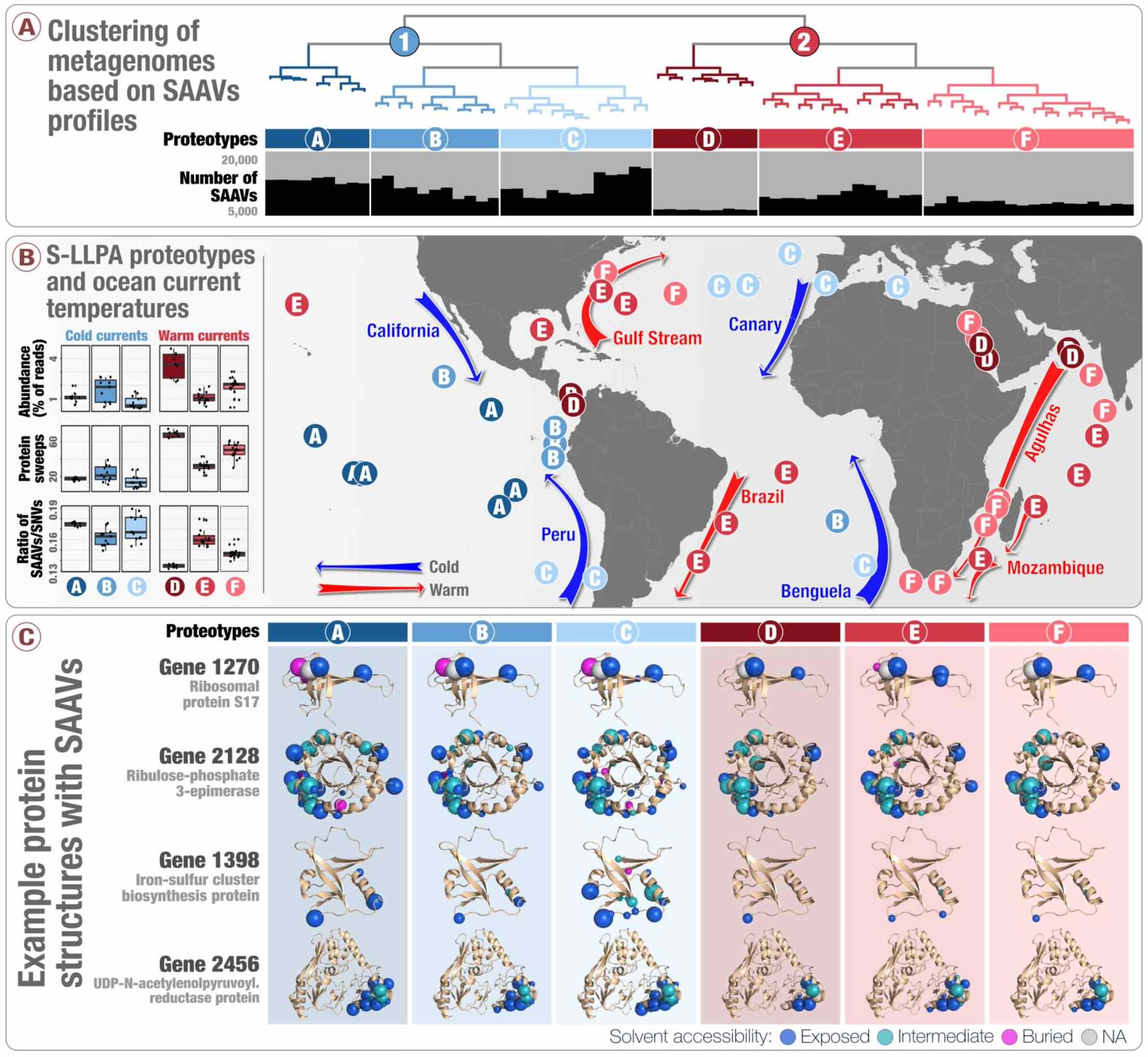
- Introduces 'single-amino acid variants' (SAAVs) and demonstrates the use of SAAVs to tease apart evolutionary processes that shape the biogeography and genomic heterogeneity within a SAR11 population through metagenomics.
- A first attempt to link population genetics and the predicted protein structures to explore in silico the intersection beetween protein biochemistry and evolutionary processes acting on an environmental microbe.
- An application of metapangenomics to define subclades of SAR11 based on gene content and ecology.
- Reproducible bioinformatics workflow is here. Reviewer criticism and our responses are also available.
2017
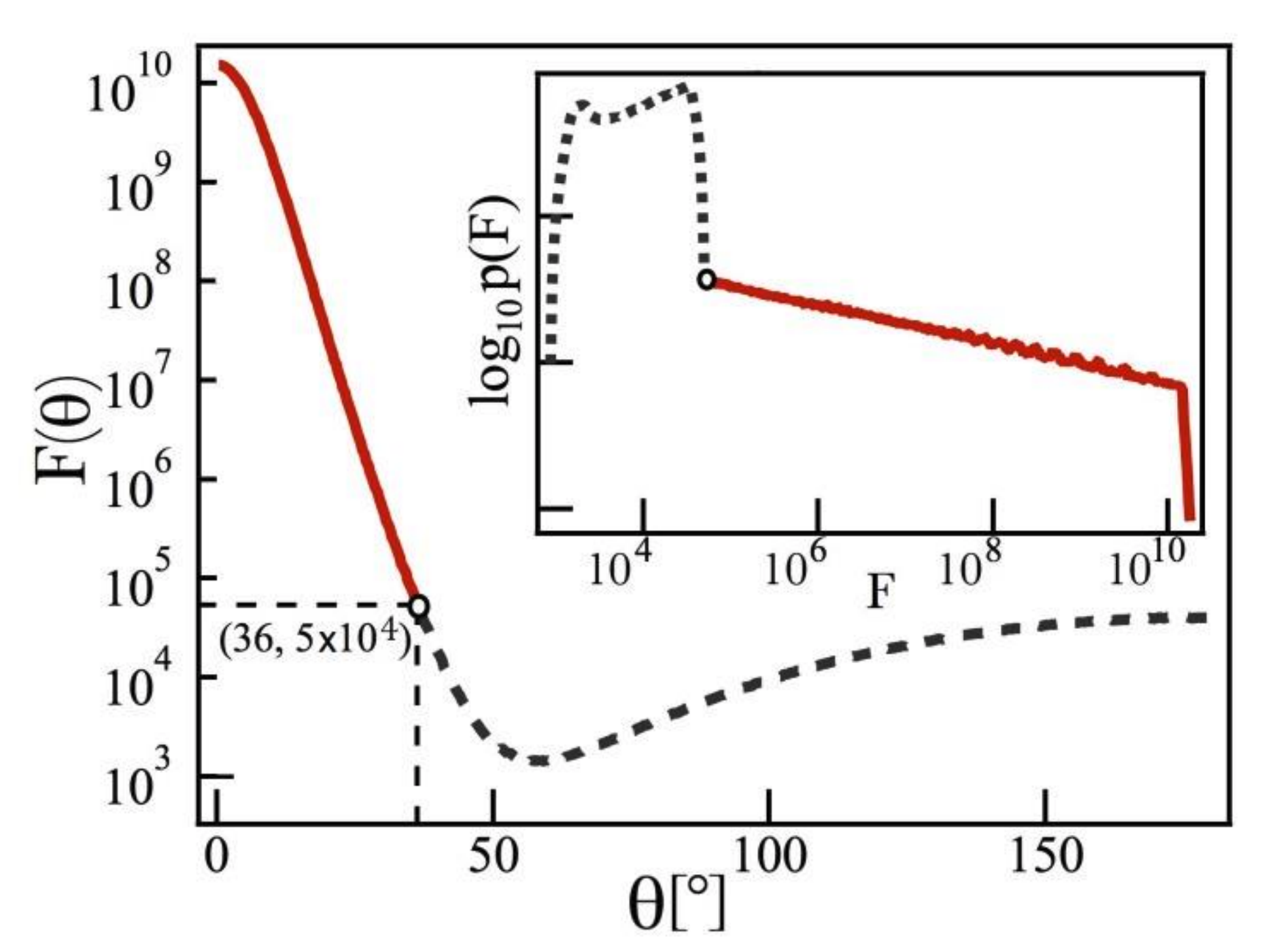
- Demonstrates power-law statistics of surface-ehanced Raman spectroscopy (SERS) hotspots can be used to assess the quality of SERS substrates.
- Extends the theory of truncated Pareto-distributed single-molecule SERS statistics to multi-hotspot substrates.
2016
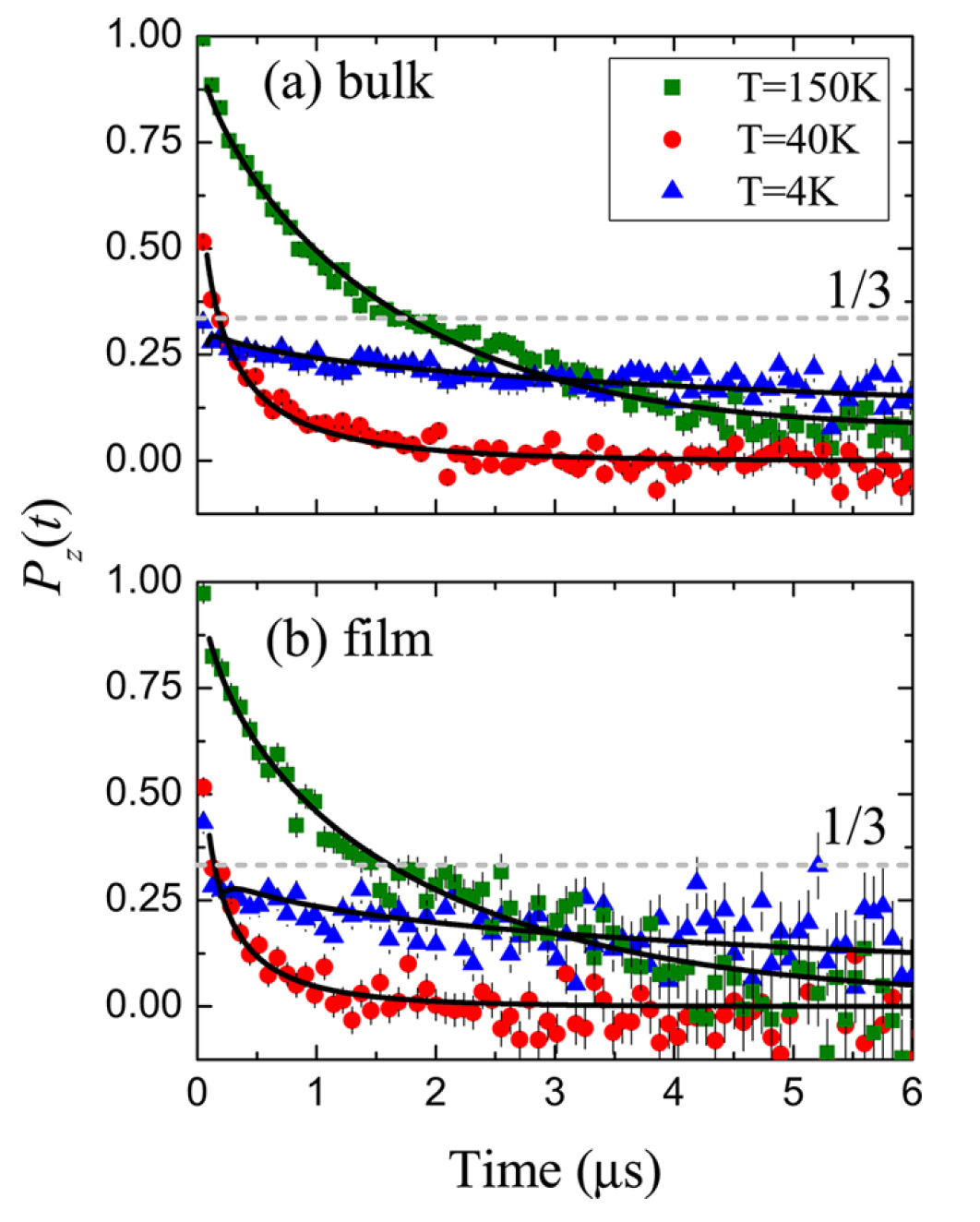
- Demonstrates an equivalence in the magnetic properties between bulk and nanofilm configurations of a single-molecule magnet (SMM) using muon spin spectroscopy
- Discovers a rare instance in which a single molecule magnet maintains its chemical structure and magnetic properties when sublimated into nanofilm, an important precursor for using SMMs for information storage.
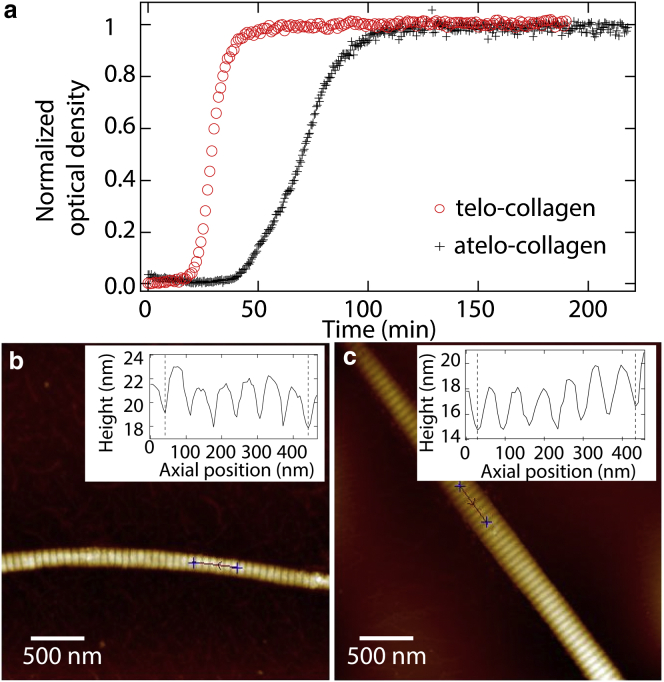
- Uses optical tweezers-based microrheology to quantify the viscoelasticity of triple-helical collagen molecules, with and without non-helical flanking regions called telopeptides, which are known to be critical for self-assembly.
- This work suggests that telopeptides facilitate transient intermolecular interactions between collagen proteins